Comprehensive Analysis of Protein Dynamics Using HiBiT Tagging: Localization, Protein Interactions, and Degradation
Simon Moe, Promega Corporation
Publication date: February 2025
Introduction
Advantages of HiBiT Technology
SMARCA2 as a Model System to Study Protein Dynamics
Validation of SMARCA2-HiBiT
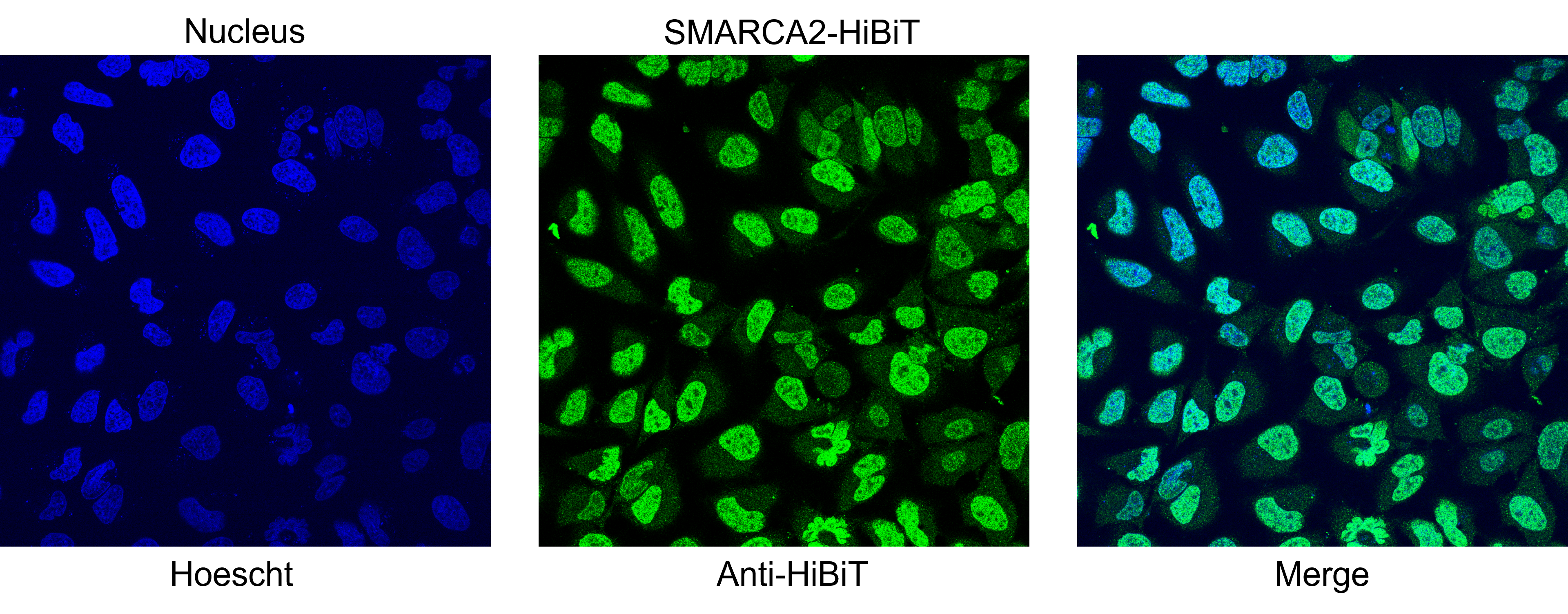
Localization of SMARCA2-HiBiT
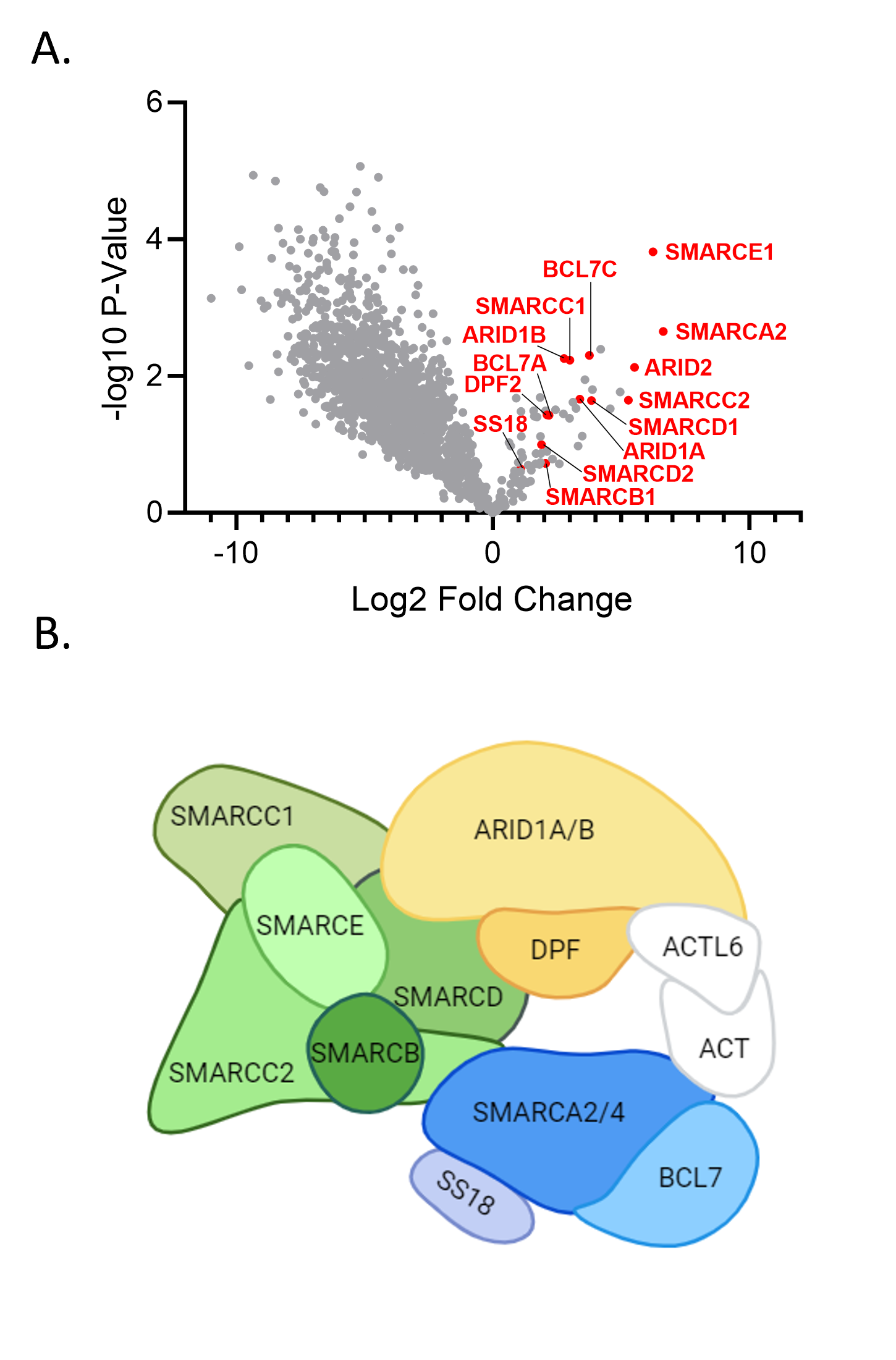
Validation of Interaction Partners
Investigations into the Protein Dynamics of SMARCA2
Using PROTACs to study Targeted Protein Degradation

Monitoring Ternary Complex Formation with NanoBRET®
Having characterized SMARCA2’s native interaction network, we set out to investigate the kinetics and mechanism of action of the ACBI1 proteolysis targeting chimera (PROTAC) that targets SMARCA2 for degradation. We leveraged NanoBRET® technology to detect ternary complex formation and ubiquitination, key steps in the mechanism of action of PROTACs. The NanoBRET® system, when combined with HiBiT tagging, provides a sensitive, real-time method to monitor these induced interactions directly in living cells.
The ternary complex forms when a PROTAC binds both a target protein and effector E3. NanoBRET® reports on complex formation by energy transfer from complemented HiBiT-LgBiT as a luminescent energy donor to a HaloTag® fluorescent acceptor when target protein and E3 are in proximity. To measure ternary complex formation, SMARCA2-HiBiT cells were co-expressed with LgBiT and HaloTag®-VHL, representing the E3 ubiquitin ligase recruited by the ACBI1 PROTAC. HaloTag® NanoBRET® 618 Ligand was added when cells were plated in assay plates. The following day cells were treated with either 1 µm ACBI1 or DMSO for 2 hours after which the NanoBRET® Nano-Glo® Substrate was added and the NanoBRET® ratio was measured on a GloMax® Discover. ACBI1-treated cells had an increase in NanoBRET® ratio compared to DMSO-treated cells, indicating the PROTAC facilitated formation of the SMARCA2-VHL complex (Figure 4), supporting the mechanism of VHL-mediated degradation of SMARCA2.
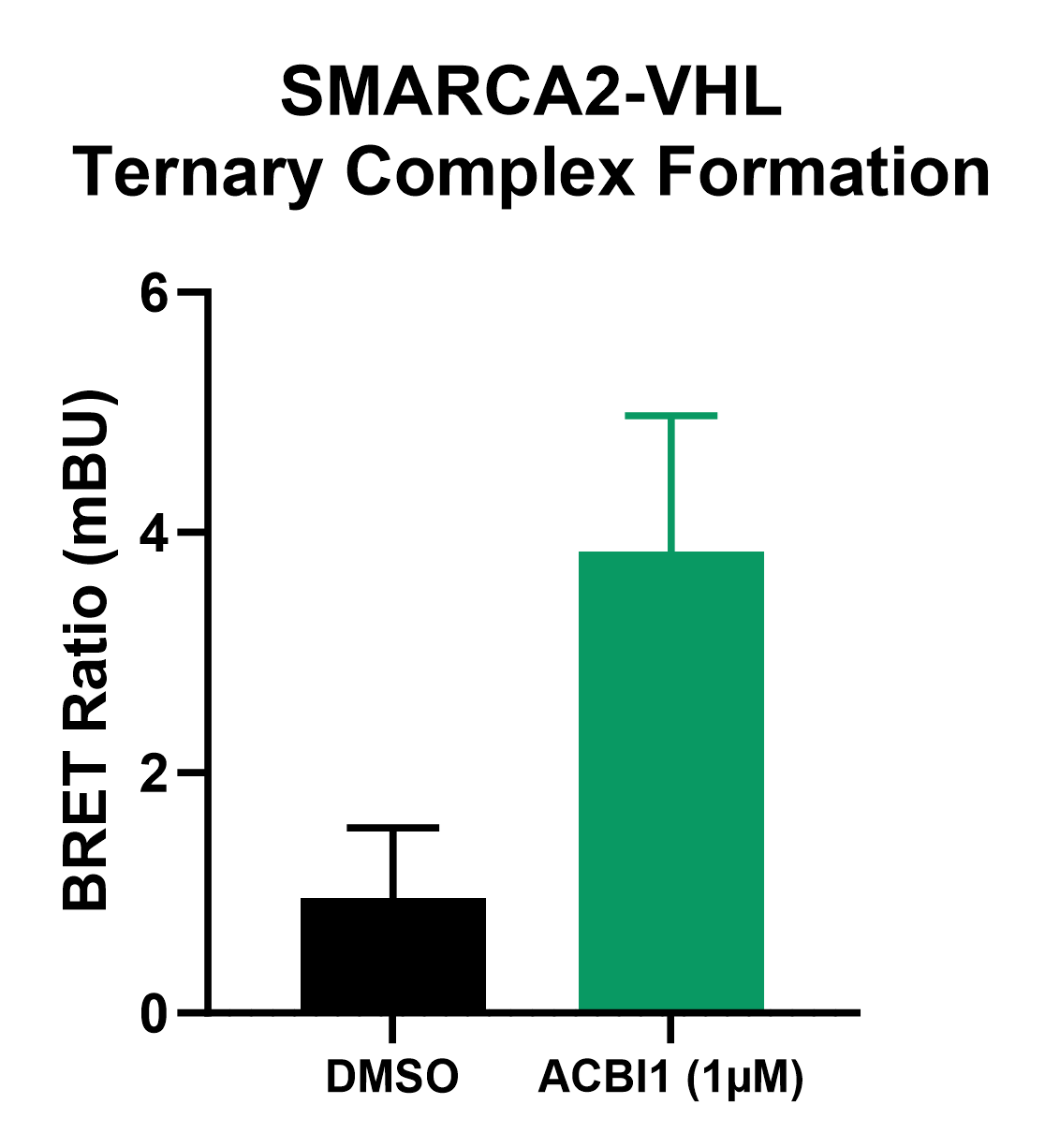
Monitoring Ubiquitination with NanoBRET®
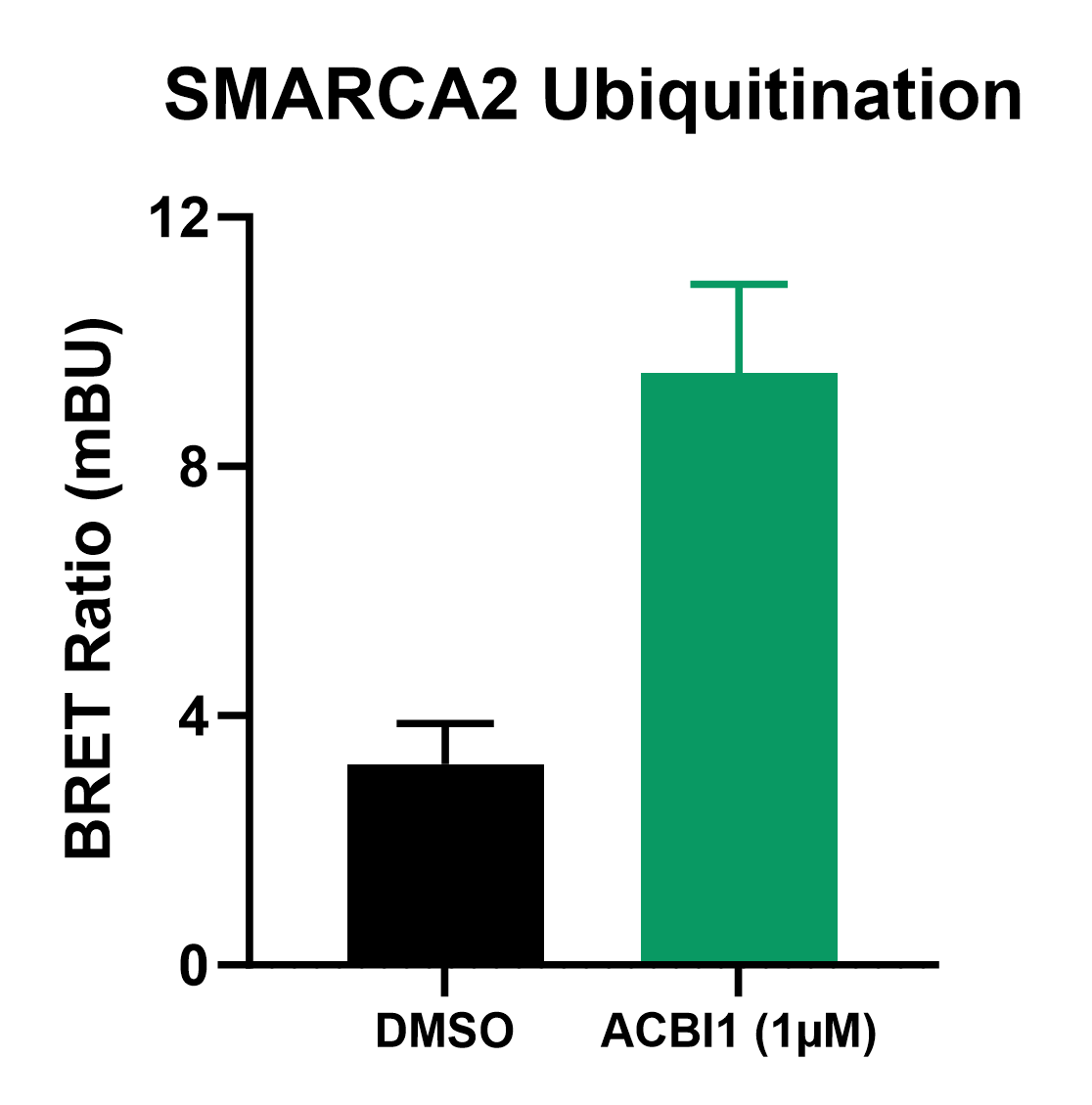
Degradation Kinetics of SMARCA2-HiBiT in Live Cells
After assessing ternary complex formation and ubiquitination, we were interested in examining the kinetics of SMARCA2 degradation within live cells. ACBI2 was chosen to induce degradation in live cells. ACBI2 is a PROTAC designed specifically to target SMARCA2, whereas ACBI1 targets both SMARCA2 and SMARCA4. Live-cell, kinetic quantification of degradation using the HiBiT tagging system offers significant advantages, including minimizing the risk of overlooking dynamic cellular responses to degrader that may be missed with end-point assays alone.
To quantify SMARCA2-HiBiT degradation, we first introduced LgBiT into live cells with the ViaScript® LgBiT mRNA Delivery System. The next day Nano-Glo® Endurazine Substrate was added to cells and given 2.5 hours to reach substrate equilibrium before the cells were treated with a dilution series of ACBI2. The luminescent signal from SMARCA2-HiBiT cells was measured over 24 hours (Figure 6). The real-time degradation profile of SMARCA2 in response to PROTAC treatment revealed rapid, potent, and near-complete degradation of SMARCA2 that persisted over time.
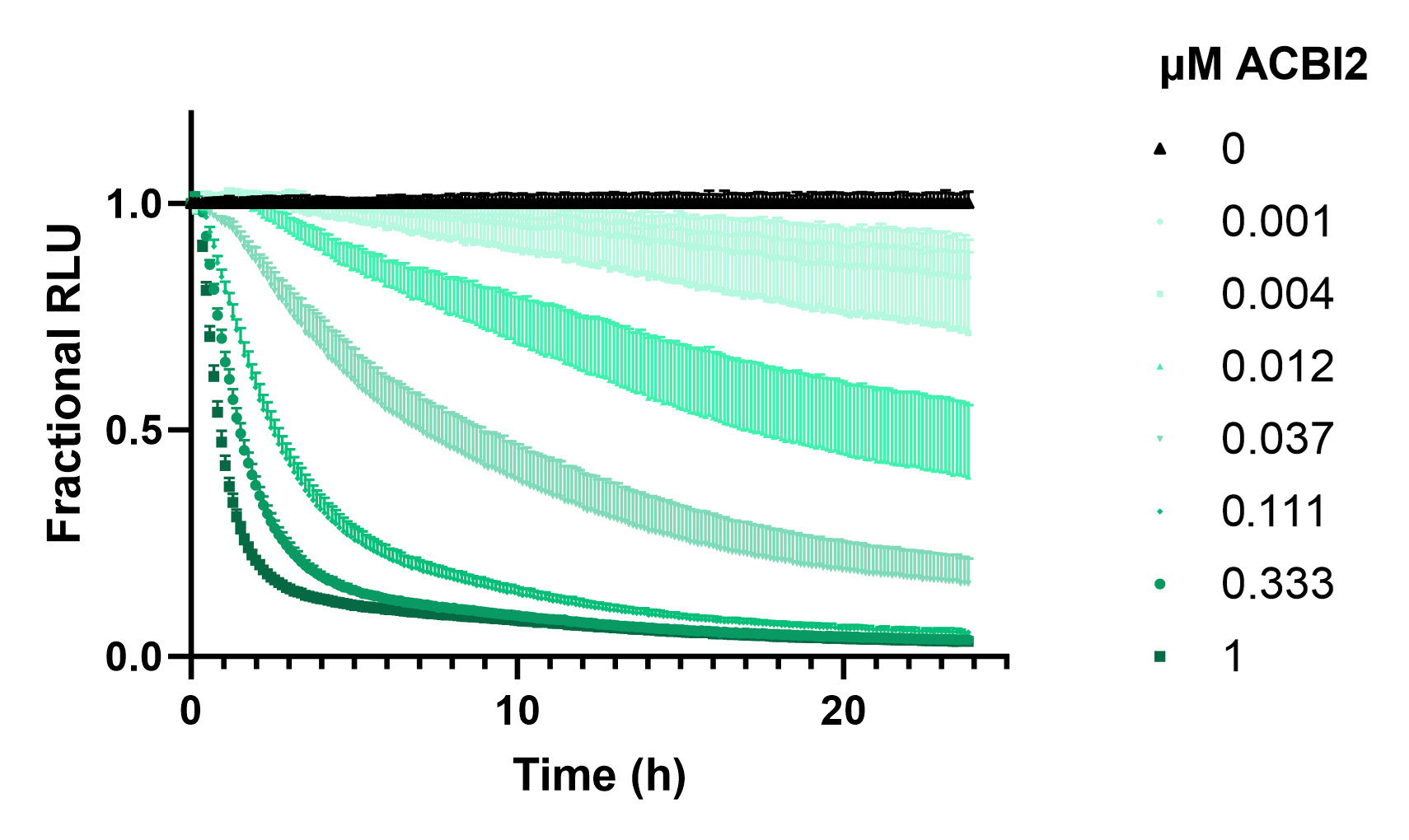
The kinetic profile provides critical insight into the action of ACBI2, and the efficiency of all mechanistic steps following from ternary complex formation to proteasomal degradation. Such data are invaluable for optimizing PROTAC-based therapeutic strategies by quantifying the relationships between concentration, time, and degradation efficacy, facilitating more informed compound ranking as well as providing deeper insights into degrader mode of action.
HiBiT technology, with its high sensitivity and non-intrusive tagging, enables precise, live-cell monitoring of protein levels in response to targeted treatments. By combining assays that elucidate the kinetics and mechanistic steps of targeted protein degraders with findings on localization and native protein interactions, we gain a comprehensive understanding of induced SMARCA2 degradation within its native cellular context. This underscores the value of HiBiT technology for detailed investigations of protein dynamics under endogenous cellular regulation.
Conclusion
The HiBiT tagging system, with its small size, high sensitivity, and compatibility with multiple detection platforms, is a versatile tool for investigating protein dynamics in their endogenous cellular environment. In this study, we demonstrated HiBiT’s utility for characterizing several aspects of SMARCA2 biology:
- Maintenance of Native Properties: SMARCA2-HiBiT was shown to localize to the nucleus, as expected for a chromatin remodeler. Additionally, IP-MS analysis revealed that SMARCA2-HiBiT interacts with expected SWI/SNF complex core proteins. The ability of HiBiT to capture these interactions illustrates its sensitivity and specificity for mapping protein-protein interactions and further exemplifies its non-disruptive nature in faithfully reporting endogenous biology.
- Characterization of Targeted Protein Degradation Mechanisms: The formation of ternary complexes and subsequent ubiquitination, assessed with NanoBRET® technology, provided insight into the mechanisms by which SMARCA2 is recruited to VHL by PROTAC ACBI1. Live-cell monitoring of degradation kinetics with ACBI2 revealed a rapid dose-dependent degradation profile, highlighting the power of HiBiT in studying dynamic processes.
We demonstrate the ability for HiBiT to capture dynamic events inSMARCA2 biology in both native and pharmacologically induced contexts, and in real time. Together, these findings highlight the versatility of HiBiT as a minimally invasive, powerful tagging system for both foundational and applied research. With its adaptability across different assay platforms, HiBiT opens new avenues for studying endogenous protein behavior, helping to unlock a deeper understanding of complex biological processes, and expanding the toolkit available for drug discovery and therapeutic development.
Citations
- Vangamudi, B., et al. (2015). The SMARCA2/4 ATPase domain surpasses the bromodomain as a drug target in SWI/SNF-mutant cancers: Insights from cDNA rescue and PFI-3 inhibitor studies. Cancer Research, 75(18), 3865–3878. https://doi.org/10.1158/0008-5472.CAN-14-3798
- Dagogo-Jack, I., et al. (2023). 713TiP A phase I study of PRT3789, a potent and selective degrader of SMARCA2 in patients with advanced or metastatic solid tumors and a SMARCA4 mutation. Annals of Oncology, 34, S493–S494. https://doi.org/10.1016/j.annonc.2023.09.1899
- Lee, E. C. Y., et al. (2024). Synthetic lethality: targeting the SMARCA2 bromodomain for degradation in SMARCA4-deficient tumors – a review of patent literature from 2019–June 2023. Expert Opinion on Therapeutic Patents, 34(4), 211–229. https://doi.org/10.1080/13543776.2024.2355258
Interested in CRISPR Knock-In Tagging?
Related Resources
Bioluminescence Imaging Solutions for Targeted Protein Degradation
Learn how the GloMax® Galaxy enables imaging the degradation of HiBiT-tagged proteins.
Targeted Protein Degradation Services
Accelerate the discovery and development of degrader molecules with our comprehensive screening and profiling services.